| |
| |
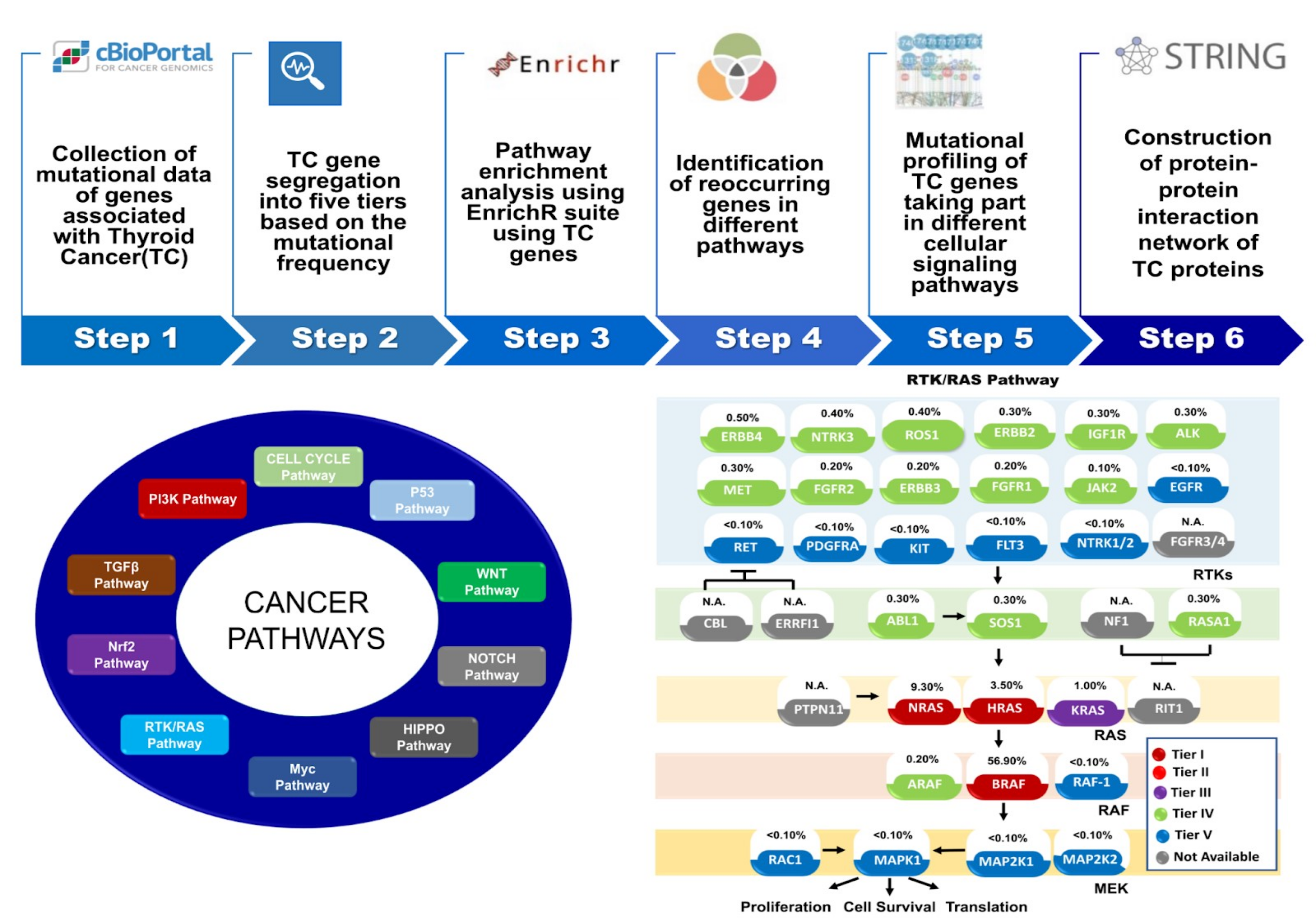 |
Oncogenomic datasets of various cancers are available in cBioPortal.
We have developed a pipeline for collection of gene mutation profiles of a particular
cancer and perform systemic analysis of pathway-based stratification for a given cancer
type and document roles of gene mutations in known cancer pathways.
| |
|
| |
 |
|
| |
| |
 |
PathBuilder is an open source software
to annotate biological information pertaining to signaling
pathways and, with minimal additional effort, to create
web-based pathway resources. PathBuilder enables annotation
of molecular events including protein-protein interactions,
enzyme-substrate relationships and protein translocation
events via manual or automatic methods. The features
of PathBuilder include automatic validation of data formats,
built-in modules for visualizing pathways, automated
import of data from other pathway resources, export of
data in several standard data exchange formats and an
application programming interface for retrieving pathway
datasets.

|
|
| |
| |
 |
|
| |
| |
 |
|
| |
| |
 |
First of its kind, a database hosting
over 25000 human proteins and disease genes. The main
objective of this database is to make available all related
fields of a protein including Interactions, Post Translational
Modifications, Substrates and other information so as
to help the scientists working in the areas of Proteomics.
This makes HPRD a reliable source for protein data. HPRD
is the first ever database to implement the standardization
protocol put forward by the Proteomics Standards Initiative
for molecular interactions (PSI-MI). The annotations
are manually done by scientists working at IOB and each
annotation undergoes multiple levels of reviews before
being made publicly available, to ensure the quality.
The database host annotations of all the known human
proteins and update them on an ongoing basis.

|
|
| |
| |
 |
A content management system, designed
for easy modifications and management of HPRD, using
web interface. The administration of annotation process
and the multi-level review carried out round the globe
is made easy by the implementation of this tool. This
user friendly tool is created using Python and Zope.

|
|
| |
| |
 |
The Plasma Proteome Database, the first
of its kind ensures a comprehensive resource for all
human plasma proteins along with their isoforms. The
database includes information pertaining to isoform specific
expression, disease, localization, post translational
modification and single nucleotide polymorphism. The
information provided in this database is through manual
annotation done by exhaustive literature research.

|
|
| |
| |
 |
TAGmapper
Is a comprehensive tool used to perform
tag-to-gene mapping. The 10 base pair sequences of each
of the SAGE tags, which are generated experimentally
in the lab, are submitted to the tool as input. The output
results in mapping the SAGE tags to their respective
genes by performing an extensive search across the ‘dbEST’ and ‘non
redundant’ database.

|
|
| |
| |
 |
ANALYSIS OF HUMAN 'X' CHROMOSOME
Involves a careful and comprehensive
analysis of Human 'X' chromosome. Using comparative genomic
approach, we have identified novel protein coding regions;
we have performed an extensive pseudogene analysis of
the 'X' chromosome and have documented alternative splicing
events. In order to help the scientific community working
on X-Linked Mental Retardation, the domain is linked
to HPRD where the user can view the annotations of all
the genes on 'X'.

|
|
| |
| |
 |
ANALYSIS OF HUMAN PROTEIN TYROSINE PHOSPHATASES FOR ALTERNATIVE
SPLICING
A thorough analysis of the protein
tyrosine phosphatases encoded by the human genome using
computational biology methods. Primary aim of the study
is to identify novel tyrosine phosphatases and novel
transcript variants for all the known protein tyrosine
phosphatases. The findings are then experimentally validated
by experts using techniques such as RT-PCRs and Northern
blots.
|
|
| |
| |

|
IOB, in collaboration with Chinnaiyan Lab, has created a microarray database whose goal is
to curate publicly available cancer microarray studies and provide data mining tools to generate biologically
relevant information in a user friendly manner. Links to various bioinformatics resources have been implemented
including Unigene, Swissprot, Biocarta, HPRD, and KEGG, among others. IOB was involved in implementing the technical
aspect for the database and Chinnaiyan Lab provided the normalized expression data. This database has been published
in January/February issue of Neoplasia.

|
|
| |

|
This database provides details of scientists and physicians involved in cancer research in India along with the information about their areas of expertise, research publications and funded grants. The main goal of the database was to foster collaborations among researchers and to provide a snapshot of ongoing research initiatives and activities in India.

|
|








